paired end sequencing read length
Modern nextgeneration sequencing platforms offer a range of read configurations such as single-read SR and paired-end PE sequencing with 75 bp per read 100 bp per read and 150 bp. The paired-end short read lengths are always 2 x 150bp 300bp.
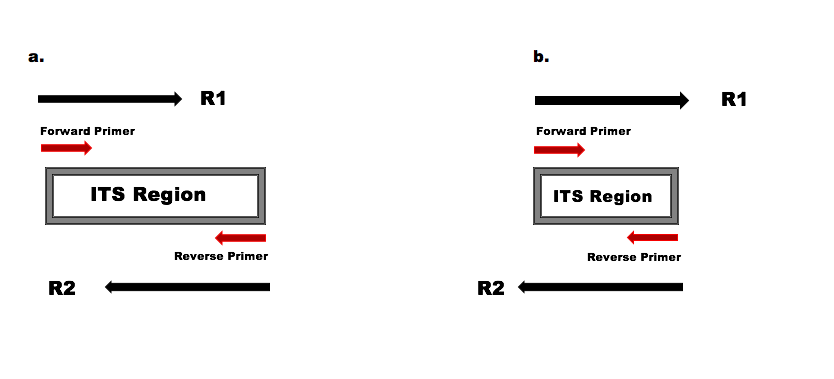
Dada2 Its Pipeline Workflow 1 8
MO 2 x 151bp.

. When using RNA-Seq to study gene expression read length is not a significant factor. Read length During sequencing it is possible to specify the number of base pairs that are read at a time. We use an Illumina MiniSeq for our short-read sequencing runs.
What matters is read counts. For example one read might consist of 50 base pairs 100 base pairs or more. The maximum distance x for a pair considered to be properly paired SAM flag 02 is calculated by solving equation Phix-musigmaxLp0 where mu is the mean sigma.
HO 2 x 151bp. However some paired-end sequencing data show the presence of a subpopulation of reads where the second read R2 has lower average qualities. Whether you align 100bp paired reads or a 50bp single.
Reading the nucleotides from two ends of a DNA fragment is called paired-end tag PET sequencing. Therefore a robust tool is needed to merge paired-end reads that exhibit varying overlap lengths because of varying target fragment lengths. When the fragment length is longer than the combined read length there.
The library prep protocols are designed to. We present the PEAR. The target DNA fragments produced by ART provide the ground truth for the merged paired-end reads.
We also generated an additional set of 150-bp long reads with a. MO 2 x 151bp. Maximum Read Length.

An Overview Of Next Generation Sequencing Technology Networks
A Genetic Algorithm For Diploid Genome Reconstruction Using Paired End Sequencing Plos One

Design Considerations Functional Genomics Ii

Paired End Vs Single End Sequencing Reads Youtube

The Variables For Ngs Experiments Coverage Read Length Multiplexing

Rapid Low Cost Assembly Of The Drosophila Melanogaster Reference Genome Using Low Coverage Long Read Sequencing Biorxiv

Denoising Of Paired End Reads Technical Support Qiime 2 Forum
Local De Novo Assembly Of Rad Paired End Contigs Using Short Sequencing Reads Plos One
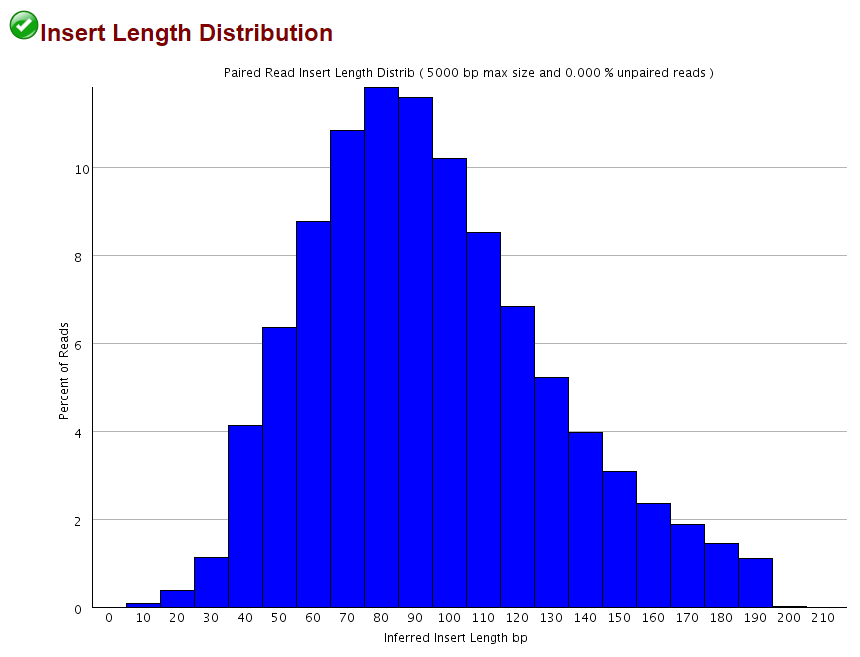
Qc Fail Sequencing Mispriming In Pbat Libraries Causes Methylation Bias And Poor Mapping Efficiencies

Assessing The Performance Of The Oxford Nanopore Technologies Minion Sciencedirect
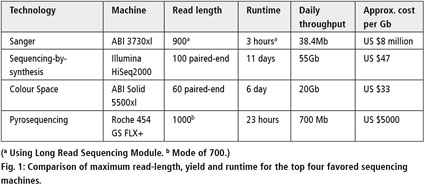
Analyzing Next Generation Sequence Data 2011 Wiley Analytical Science

Interpreting Color By Insert Size Integrative Genomics Viewer
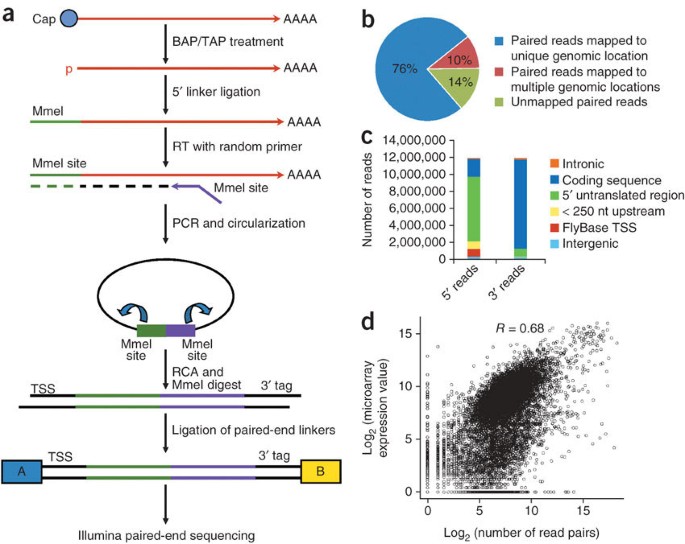
A Paired End Sequencing Strategy To Map The Complex Landscape Of Transcription Initiation Nature Methods

What Is Mate Pair Sequencing For
A Comprehensive Evaluation Of Single End Sequencing Data Analyses For Environmental Microbiome Research Springerlink
Direct Chloroplast Sequencing Comparison Of Sequencing Platforms And Analysis Tools For Whole Chloroplast Barcoding Plos One


